10.2 Modern genetics
Genetic information
Genetic information is encoded in DNA
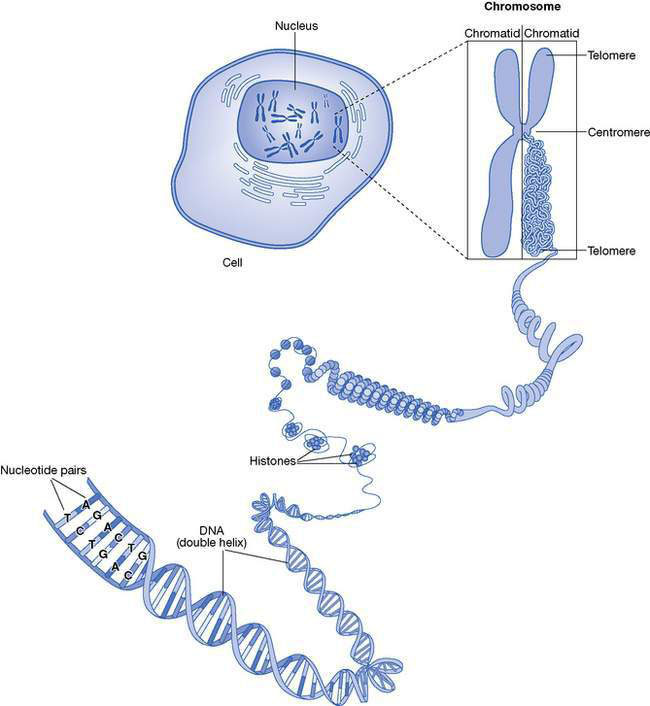
Deoxyribonucleic acid (DNA) is a complex molecule located within the nucleus of every cell of a body (Fig. 10.2.1.) It is the biological ‘library’ of the genetic information needed for a fertilized egg to develop into a complex and functional organism (a person) comprised of a wide variety of cell and tissue types.
• DNA is a linear molecule comprised of subunits called nucleotides.
• There are four different nucleotides; adenosine (A), thymidine (T), cytosine (C) and guanidine (G).
• Each nucleotide is linked to its immediate neighbour by a sugar molecule, deoxyribose, forming a long chain.
• Two chains of nucleotides are linked together by hydrogen bonds.
• Adenosine in one chain pairs with thymidine in the other chain, and cytosine pairs with guanidine.
• The hydrogen bonds between the A–T and C–G pairs are like the rungs on a ladder.
• The entire ladder is twisted along its length into a conformation that is often called a ‘double helix’.
• The entire DNA sequence is called the human genome and is encoded by approximately three billion (3 × 109) nucleotides.
Nuclear DNA forms tightly coiled lengths of DNA called chromosomes
• Human somatic (‘body’) cells have two copies of the nuclear genome, one copy inherited from each parent; this DNA is packaged into 46 chromosomes (23 pairs).
• Sperm and ova (gametes) have a single copy of the nuclear genome packaged as 23 unpaired chromosomes.
• Fertilization of an ovum by a sperm restores the usual amount of nuclear DNA in a somatic cell; 46 chromosomes; 23 pairs.
• In a karyotype, chromosome pairs are arranged in a standard format based on their size, banding pattern and centromere position; the first 22 pairs are called autosomes; the 23rd pair are the two sex chromosomes.
• In females there are two equivalent long sex chromosomes called X chromosomes.
• In males there is one long X chromosome and a small Y chromosome.
One X chromosome is inactivated in females
• At the time of conception both X chromosomes in a female conceptus are active.
• After a few cell divisions, one X chromosome in each cell is inactivated by methylation; the same X chromosome remains inactive in all daughter cells derived from that ancestral cell.
• The result is that both males and females have only one active X chromosome in each cell.
• On average, each of the two X chromosomes will be active in half of the cells of a female’s body.
• Because X inactivation is initiated when the conceptus consists of a small number of cells, by chance approximately 10% of females have the same X chromosomes active in 90% or more of cells (skewed X inactivation).
Mitochondrial DNA is a small, circular, double-stranded DNA
• mtDNA is a double-stranded loop, 16 569 nucleotides long, that is joined end-to-end in a circle.
• mtDNA encodes 13 proteins involved in oxidative phosphorylation, as well as 24 non-coding ribonucleic acids (RNAs) important for normal mitochondrial function.
• Each mitochondrion has 5 to 10 copies of mtDNA and, depending on the type of tissue, a single somatic cell may contain as many as 1000 mitochondria.
• Tissues that are highly dependent on oxidative phosphorylation (e.g. neurons) have more mitochondria per cell than tissues that are less dependent (e.g. some epithelial cells).
• Mitochondria and mtDNA are copied independently of the process of copying nuclear DNA.
• Mitochondria are maternally inherited – all the mitochondria in an individual’s cells are derived from the original ovum.
• The exclusive maternal inheritance of the mitochondrial DNA is in marked contrast to the bi-parental inheritance of nuclear DNA.
The function of our genome: DNA, genes and proteins
Protein-coding genes
• A gene consists of a sequence of nucleotides.
• Some genes are encoded by several hundreds of nucleotides and others by many thousands.
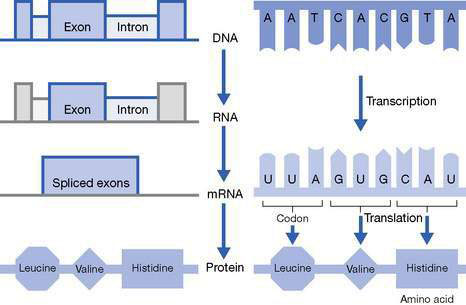
• The DNA sequence of most nuclear genes is broken up into coding modules (exons), which are separated from each other by regions of non-coding DNA (introns) (Fig. 10.2.2).
• Mitochondrial genes do not have introns.
• Specific DNA sequences define the beginning and the end of a gene.
• Exons are usually a few hundred nucleotides long.
• Introns may be many thousands of nucleotides in length.
• Introns have regulatory and evolutionary roles that are poorly understood.
Non-protein-coding genes
A unit of DNA sequence that encodes a functional, non-coding RNA transcript is also called a gene.
The DNA sequence of a gene is transcribed into RNA
To access the genetic information encoded in a gene, the enzyme RNA polymerase uses the gene’s nucleotide sequence as a template to synthesize a molecule of RNA, a process called transcription (see Fig. 10.2.2).
• RNA has a structure that is similar to DNA, except it is usually single-stranded and uracil (U) substitutes for thymidine.
• DNA is transcribed to a primary RNA transcript; a series of processing reactions follow, including removal of introns, producing a shorter, mature RNA transcript.
• There are many different types of mature RNA that have a range of different functions, some unknown.
• Protein-coding genes encode a special form of mature RNA, messenger RNA (mRNA), which serves as a template for synthesis of a protein polypeptide.
mRNA is translated into protein via a triplet nucleotide code
• mRNA is translated (decoded) to make a protein polypeptide at ribosomes.
• The information encoded in the mRNA sequence is interpreted via a triplet nucleotide code (a codon), which determines the exact amino acid sequence of the protein (see Fig. 10.2.2).
• There are also specific stop codons that indicate the end of a gene.
Our understanding of the function of DNA is changing
• About 5% of the human genome sequence has been highly conserved by evolution, and most of this DNA is functionally important.
• Around one-fifth of this conserved sequence encodes proteins (protein-coding DNA).
• The other four-fifths of this conserved sequence is non-protein-coding DNA which encodes functionally important RNA molecules that are not translated into proteins (i.e. non-coding RNA transcripts).
• Although the remaining 95% of the human genome sequence is less conserved, it is estimated that around 85% is transcribed to non-coding RNA transcripts.
Gene regulation
The regulation of human gene expression is incompletely understood
• Methyl groups interfere with the binding of molecules to the regulatory sequence of a gene.
• The net effect of this methylation is inactivation of the gene.
• Methyl groups may also be removed, activating a gene.
• Methylation provides a means for varying gene activity in normal cells.
• Some genes are inactivated in cells of a specific tissue type(s), resulting in tissue-specific expression of the gene.
• Some genes are inactivated at certain times during development leading to developmental stage-dependent expression.
• X chromosome inactivation in females is an example of gene inactivation due to methylation.
• Imprinting is the selective inactivation of a gene according to the sex of the parent who passed on that gene.
• Imprinting is a normal process and results in only one active copy of the gene in a cell.
• Inactivation occurs because of methylation of a regulatory region of the gene.
• Some imprinted genes are selectively inactivated when transmitted by sperm and selectively activated when transmitted by an ovum (they are paternally imprinted).
• Other genes demonstrate the reciprocal pattern and are selectively inactivated when transmitted by an ovum and selectively activated when transmitted by a sperm (they are maternally imprinted).
• Normally the methylation ‘imprint’ is removed in sperm and ova so that inactivation of the imprinted gene is always determined by the sex of the parent of origin.
• Imprinting is an example of an epigenetic mechanism; a heritable change in gene expression that does not occur because of a change in DNA sequence.
• Failure to imprint a gene results in there being two active copies of the gene, producing a ‘functional duplication’ of that gene.
• Inappropriate imprinting of a gene results in there being two inactive copies of the gene, producing a ‘functional deletion’ of that gene.
• Examples of imprinting disorders include Prader–Willi syndrome, Angelman syndrome, Russell–Silver syndrome and Beckwith–Wiedermann syndrome.
Uniparental disomy
When the paternal copy of the gene is normally imprinted:
• if both copies of the gene are inherited from the father there are zero active copies of the gene instead of the normal single active copy and a ‘functional deletion’
• if both copies of the gene are inherited from the mother there are two active copies of the gene and a ‘functional duplication’.
When the maternal copy of the gene is normally imprinted:
Genes and networks
Cystic fibrosis (CF) is a common autosomal recessive disorder:
• It affects approximately 1 in 2500 Caucasian children.
• 1 in 25 healthy Caucasian adults are carriers of a mutation that can cause CF.
• More than 2000 different mutations have been identified in the CF gene (genetic heterogeneity).
• One mutation (deltaF508) accounts for about 80% of all mutations; other mutations are much less common.
• Without treatment, children with classical CF usually die during infancy.
• With treatment, at least 50% of these children are alive in their mid-40s.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree