Human Cytogenetics
Sarah T. South and John C. Carey
The field of clinical cytogenetics and the description of syndromes caused by gross chromosomal abnormalities laid the foundation for defining and delineating malformation syndromes. Chromosomal abnormalities are detected in approximately 1 in 110 newborns and are the common most single cause of mental retardation or developmental delay.1,2 The common pediatric indications for a chromosome analysis include growth retardation, neurologic impairment, neuropsychological dysfunction, ambiguous genitalia, or multiple congenital anomalies. Clinical cytogenetics also, in part, laid the foundation of the field of dysmorphology. This chapter provides the principles of human cytogenetics.
BASICS OF A CHROMOSOME ANALYSIS
Cytogenetics is a whole genome analysis involving the examination of chromosomes from a tissue of interest to identify large-scale genomic alterations. This occurs through the microscopic examination of chromosomes arrested during the metaphase stage of cell division. The chromosomes are treated with enzymes and chemicals to produce characteristic light and dark patterns, called bands, along the arms of the chromosomes. Each of the 46 chromosomes can then be identified individually and organized into a karyogram  and described as a karyotype (the nomenclature used to describe the results of the chromosome analysis, described in more detail below).
and described as a karyotype (the nomenclature used to describe the results of the chromosome analysis, described in more detail below).
The benefits of a chromosome analysis include visualization of the entire genome on a cell-by-cell basis, which allows for the nonselective identification of large-scale alterations in genome structure, as well as detection of mosaicism (the presence of two or more distinct cell populations within an individual). The limitations of this technology include a limit to the size of a genomic abnormality that can be detected. This limit of resolution is about 10 megabases (Mb) but varies according to the region of the genome in which the abnormality occurs and the quality of the chromosome preparations, because abnormalities will be detected only if they alter the banding pattern. Another limitation of a standard chromosome analysis is the need for an actively growing source of cells. At the time of sample acquisition, the majority of cells will not be in metaphase, and therefore, must be cultured, often with chemicals that increase the number of cells in metaphase at the time the cells are harvested and prepared for analysis. As a result, cells that have been fixed or are no longer viable cannot be analyzed with this technology.
 NOMENCLATURE
NOMENCLATURE
Normally, each human has 46 chromosomes that are distributed in 23 pairs, 22 pairs of autosomes and 1 pair of sex chromosomes (XX in females and XY in males). Thus, each individual has two copies of each chromosome (ie, diploid). A normal chromosome constitution is termed the euploid state, whereas an abnormal chromosome complement is called aneuploidy. The autosomes are numbered 1 to 22, with the numbers assigned in descending order of length, size, and centromere position of each chromosome pair, although chromosome 21 is actually smaller than chromosome 22. The position of the centromere varies among chromosomes and is classified into three categories: metacentric chromosomes have centromeres in the middle, submetacentric chromosomes display centromeres closer to one end, and acrocentric chromosomes have centromeres at the end of the chromosome. For example, the chromosomes 13, 14, 15, 21, and 22 are referred to as acrocentric.
The centromere (cen) divides a chromosome into a short (p) arm and a long (q) arm. Each arm ends in a terminus (ter). Thus, the end of the short arm is called pter. The arm of each chromosome is divided into regions, these regions are divided into bands, and bands into subbands. When a karyotype is reported, the total number of chromosomes is presented first, followed by the sex chromosomes’ constitution, and then any numerical and structural anomalies are indicated to the level of the subbands. The International System for Human Cytogenetic Nomenclature (ISCN) 2005 provides additional information.3 Some examples of normal and abnormal karyotype reports are given in Table 173-1.
CLASSIFICATION OF CHROMOSOME ABNORMALITIES
The common types of chromosome abnormalities that are detectable with a standard chromosome analysis include the loss or gain of an entire chromosome deletions or duplications that alter the banding pattern within a chromosome inversions of material within a chromosome and translocations of material between two chromosomes (eFig. 173.2  ). Reciprocal translocations refer to the exchange of segments between nonhomologous chromosomes (eg, between chromosomes 9 and 22). Robertsonian translocations involve the fusion of two acrocentric chromosomes in which the short arms of each chromosome are lost in these rearrangements. These acrocentric short arms contain redundant copies of ribosomal RNA genes, and thus their loss is of no clinical significance. Translocations can be either balanced, with no loss or gain of genetic material, or unbalanced, with chromosome material both changed in its position and either lost or gained. In the majority of cases, unbalanced translocations arise in an individual because of the unbalanced segregation of chromosomes from a parent that carries a balanced translocation (eFig. 173.3
). Reciprocal translocations refer to the exchange of segments between nonhomologous chromosomes (eg, between chromosomes 9 and 22). Robertsonian translocations involve the fusion of two acrocentric chromosomes in which the short arms of each chromosome are lost in these rearrangements. These acrocentric short arms contain redundant copies of ribosomal RNA genes, and thus their loss is of no clinical significance. Translocations can be either balanced, with no loss or gain of genetic material, or unbalanced, with chromosome material both changed in its position and either lost or gained. In the majority of cases, unbalanced translocations arise in an individual because of the unbalanced segregation of chromosomes from a parent that carries a balanced translocation (eFig. 173.3  ).
).
MECHANISM OF FORMATION OF ABNORMAL CHROMOSOMES AND CLINICAL CONSEQUENCE
Whole chromosome loss or gain occurs primarily through nondisjunction in either the first or second meiotic division. The majority of cases are associated with advanced maternal age and a nondisjunction event in the first meiotic division. However, sex chromosome aneuploidy, as seen in Turner and Klinefelter syndromes, does not show an association with advanced maternal age. The most common clinical consequences of whole chromosome aneuploidy in live births include trisomy 21 (Down syndrome), trisomy 13 (Patau syndrome), and trisomy 18 (Edward syndrome), along with the previously mentioned sex chromosome aneuplodies in Turner syndrome, the majority of cases in which there is only one X chromosome and no other sex chromosome, and Klinefelter syndrome, which is the result of an additional X chromosome in an otherwise male chromosome complement. Loss or gain of the other chromosomes is usually not compatible with a live birth.
Interstitial duplications and deletions, inversions, and translocations primarily involve nonallelic recombination between low copy repeats or segmental duplications within the genome. 
Balanced translocations occur in approximately 1 in 500 individuals. They are often associated with a normal phenotype in the carrier, but can be associated with a clinical consequence if the translocation disrupts a gene or a regulatory element. There is also a reproductive consequence for carriers of balanced translocations as independent segregation of genetically unbalanced gamete.  Therefore, balanced translocation carriers have a higher risk for infertility, a higher miscarriage rate, and a greater risk for the birth of an abnormal child with a genetic imbalance. The exact risk for each of these outcomes correlates with the viability of the genetic imbalance. In general, the larger genetic imbalances are associated with greater risk for infertility and miscarriage; the smaller genetic imbalances have a greater risk for the birth of an abnormal child. When counseling a family with multiple carriers of a balanced translocation, a thorough family history is very valuable in accessing reproductive risk.
Therefore, balanced translocation carriers have a higher risk for infertility, a higher miscarriage rate, and a greater risk for the birth of an abnormal child with a genetic imbalance. The exact risk for each of these outcomes correlates with the viability of the genetic imbalance. In general, the larger genetic imbalances are associated with greater risk for infertility and miscarriage; the smaller genetic imbalances have a greater risk for the birth of an abnormal child. When counseling a family with multiple carriers of a balanced translocation, a thorough family history is very valuable in accessing reproductive risk.
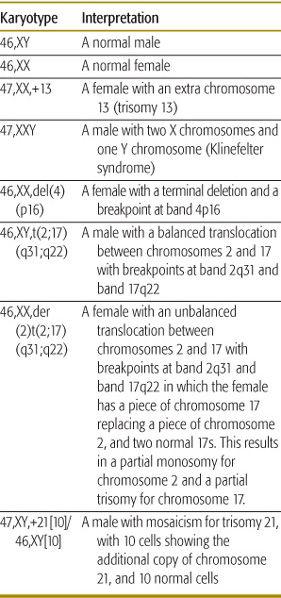
Table 173-1. Examples of Karyotypes with Interpretation
Similar to balanced translocations, balanced inversions may also disrupt genes and regulatory elements, or may be associated with no clinical phenotype in the carrier. Inversions also have reproductive consequences due to an increased risk of producing genetically unbalanced gametes as a result of the complicated structures that form to allow for homologous pairing during meiosis. 
Ring chromosomes are formed when a chromosome undergoes breaks on both sides of the centromere and the broken ends of the chromosome reunite. A marker chromosome is a structurally abnormal chromosome whose origin cannot be identified, usually because the marker chromosome is either too small or too rearranged and the banding pattern is ambiguous. Molecular cytogenetic techniques are often necessary to allow identification of marker chromosomes.
MOLECULAR CYTOGENETIC TECHNIQUES
In contrast to standard cytogenetic techniques, molecular cytogenetic techniques analyze smaller regions looking for both imbalances and rearrangements primarily through fluorescence in situ hybridization (FISH) and comparative genomic hybridization microarray (array CGH).
FISH was first described in 1986 and involves labeling DNA with a fluorescent molecule and then hybridizing this probe to the patient’s chromosomes. If there is a deletion of this DNA in the patient, there will be a failure of the probe to hybridize to the deleted chromosome. If there is a duplication of this DNA in the patient, then there will be an increase in the number of probe signals that have hybridized to the patient’s chromosomes.
The benefits of FISH include the ability to turn almost any region of the human genome into a probe; however, the majority of FISH probes for clinical use need to be at least 100 kB in size. Nevertheless, this is still a 100-fold improvement in the resolution of FISH compared with a whole chromosome analysis by traditional banding methods. Furthermore, chromosomes do not need to be in metaphase, but may be in interphase. Therefore, archived and nonviable tissue can also be analyzed. The major limitation of FISH is that it is not a whole genome analysis. FISH analyzes only the region of the genome that is complementary to the DNA probe. Therefore, only deletions, duplications, insertions, or translocations that alter the region of DNA complementary to the probe are identified.
A common example of FISH in a pediatric setting is the identification of microdeletion syndromes that cannot be seen by a standard chromosome analysis. As an example, Wolf-Hirschhorn syndrome is the result of deletions of a critical region on chromosome 4. The deletion may be as large as 20 mB and seen by chromosome banding or may be as small as 2 mB with two normal-appearing chromosome 4s by banding. However, the DNA within the critical region on chromosome 4 can be turned into a FISH probe that can then be used to identify a deletion of this region within any patient with Wolf-Hirschhorn syndrome (eFig. 173.6  ).
).
Because interphase cells from the patient can also be used to determine the presence or absence of DNA complementary to the FISH probe, culturing of the patient sample is not always necessary. This allows for a more rapid prenatal and neonatal detection of common aneuploidies. Uncultured amniocytes or leukocytes can be hybridized to FISH probes complementary to a region of chromosome 21. The detection of three copies of the chromosome 21 locus by FISH is then suggestive of trisomy 21  . This type of prenatal or neonatal interphase FISH analysis should always be followed by a standard chromosome analysis to determine the mechanism leading to additional chromosome 21 material, as this may be caused either by nondisjunction with a relatively low recurrence risk or by an unbalanced translocation with an associated higher recurrence risk.
. This type of prenatal or neonatal interphase FISH analysis should always be followed by a standard chromosome analysis to determine the mechanism leading to additional chromosome 21 material, as this may be caused either by nondisjunction with a relatively low recurrence risk or by an unbalanced translocation with an associated higher recurrence risk.
Array CGH is one of the newest technologies developed for the detection of a chromosome imbalance. This assay involves genomic DNA isolation from a test sample and a control sample, labeling of the two DNA samples with different fluorochromes, and cohybridization of the two differentially labeled DNAs to target DNAs, which are segments of the human genome spotted on a glass slide. The target DNA can be either large insert clones that are approximately 50 to 200 kb in size, or small oligonucleotide sequences, usually about 60 base pairs. The target DNA can also be from any region of the human genome, although for technical and interpretive reasons the target DNA used for arrays ideally represents unique DNA regions that are not repetitive in nature. In addition, these target DNAs can be spaced throughout the genome to provide a whole genome analysis or be concentrated in regions of the genome already associated with a genomic disorder.
If the amount of test DNA is equivalent to the amount of control DNA, then equal hybridization of the two labeled populations to the target DNA on the slide will occur. However, if the test sample has a deletion of a particular target DNA segment, then relatively more of the control DNA will hybridize to that particular target DNA on the slide. Conversely, if the test sample has a gain of a particular target DNA segment, then relatively more of the test DNA will hybridize to the target DNA on the slide. By quantifying the amount of differentially labeled DNA hybridized to each target on the slide, gains and losses in the test sample relative to the control DNA are identified. As a control, each target DNA is spotted multiple times on the slide and many laboratories also repeat the assay with the two fluorochromes reversed, known as a dye-swap. The ratios of the two fluorochromes at each target spot, from both assays of the dye-swap, are then plotted, and a true imbalance will show a reciprocal deviation at the target site. Abnormalities are often confirmed through an alternative method, such as FISH  .
.
Array CGH has proven very useful for identification of constitutional chromosomal imbalances in patients with developmental delays and detects clinically relevant imbalances in 6% to 17% of cases that have had a normal standard chromosome-banding analysis.5-7 However, a challenge of this new technology is its ability to detect a DNA copy number change of unknown clinical significance. This challenge is not unique to this new technology, but rather has existed in many new technologies. For example, analysis of single genes for diagnostic purposes may uncover a sequence change of unknown clinical significance. Further investigations may allow this sequence change to be subsequently classified as either disease causing or benign, whereas other alterations remain in the category of unknown clinical significance. A common method for determination of the clinical relevance of a DNA change, be it at the nucleotide or chromosome level, is to determine if the change is de novo or inherited from a normal parent and whether or not the change is found in normal individuals within the population.
Although very useful for detection of genetic imbalances, it should be noted that array CGH technology will not detect balanced rearrangements and will not detect gain or loss of DNA that is not spotted onto the microarray slide. Additionally, changes in the genomic architecture that may accompany a genetic imbalance are not identified by array CGH and these changes are often helpful in determining the mechanism leading to the loss or gain of genetic material. Furthermore, the technology may be limited in its ability to detect low levels of mosiacism, because the patient DNA comes from a pool of millions of cells. Therefore, a standard chromosome analysis, and occasionally FISH, is still warranted when an array CGH analysis is normal or as part of the complete characterization of an abnormality.
Although this discussion has primarily focused on the use of a chromosome analysis in the context of a pediatric population, chromosome analysis plays an important role in the prenatal and oncology fields.
As nondisjunction is often associated with advanced maternal age, this is one common indication for a prenatal analysis using either amniocytes or chorionic villi. Other common indications for a prenatal analysis include an abnormal serum screen, either in the first or second trimester; an abnormal ultrasound; a family history or a previous pregnancy with a chromosome abnormality; or a family history of an X-linked recessive disorder to determine the gender of the fetus and whether subsequent testing is warranted.
With chromosome aberrations found in approximately 40% of unselected spontaneous abortions, chromosome anomalies are the most common single cause of a pregnancy loss, particularly in the first trimester. Therefore, a chromosome analysis on either placental villi or fetal tissue from a miscarriage can frequently provide an explanation for the loss.
An oncology-related chromosome analysis can provide diagnostic information. For example, the diagnosis of chronic myelogenous leukemia requires the identification of the translocation between chromosome 9 and 22, which results in the fusion of the BCR and ABL1 genes.
An oncology-related chromosome analysis can also provide prognostic information. For example, precursor B-cell acute lymphoblastic leukemia (B-cell ALL) can be diagnosed based on numerous clinical and laboratory findings, yet certain chromosome rearrangements provide unique prognostic information, and therefore guide subsequent treatment options. Identification of a translocation involving the MLL gene on chromosome 11 in B-cell ALL is associated with a very unfavorable prognosis and indicates more aggressive therapy whereas identification of hyperdiploidy with a gain of chromosomes 4, 10, 17, and 21 is associated with a much more favorable prognosis and less aggressive therapy.
Chromosome analysis of bone marrow or tumor tissue can also monitor for secondary changes that signal disease progression, such as the acquisition of an additional copy of the abnormal chromosome 22 in chronic myelogenous leukemia (CML); thus, it can also monitor the effectiveness of therapy, as the chromosomally abnormal cells should disappear if the treatment is working. However, as patients undergo therapy for a current malignancy, it is also possible for that patient to develop a secondary, or therapy-related, leukemia and a chromosome analysis that identifies new abnormalities during or posttreatment can be one of the earliest indications of a secondary leukemia.
REFERENCES
See references on DVD.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


