Background
An evaluation for heritable thrombophilias is recommended in the evaluation of stillbirth. However, the association between thrombophilias and stillbirth remains uncertain.
Objective
We sought to assess the association between maternal and fetal/placental heritable thrombophilias and stillbirth in a population-based, case-control study in a geographically, racially, and ethnically diverse population.
Study Design
We conducted secondary analysis of data from the Stillbirth Collaborative Research Network, a population-based case-control study of stillbirth. Testing for factor V Leiden, prothrombin G20210A, methylene tetrahydrofolate reductase C677T and A1298C, and plasminogen activating inhibitor (PAI)-1 4G/5G mutations was done on maternal and fetal (or placental) DNA from singleton pregnancies. Data analyses were weighted for oversampling and other aspects of the design. Odds ratios (OR) were generated from univariate models regressing stillbirth/live birth status on each thrombophilia marker.
Results
Results were available for ≥1 marker in 488 stillbirths and 1342 live birth mothers and 405 stillbirths and 990 live birth fetuses. There was an increased odds of stillbirth for maternal homozygous factor V Leiden mutation (2/488; 0.4% vs 1/1380; 0.0046%; OR, 87.44; 95% confidence interval, 7.88–970.92). However, there were no significant differences in the odds of stillbirth for any other maternal thrombophilia, even after stratified analyses. Fetal 4G/4G PAI-1 (OR, 0.63; 95% confidence interval, 0.43–0.91) was associated with decreased odds of stillbirth. Other fetal thrombophilias were similar among groups.
Conclusion
Most maternal and fetal thrombophilias were not associated with stillbirth. Maternal factor V Leiden was weakly associated with stillbirth, and the fetal PAI-1 4G/4G polymorphism was associated with live birth. Our data do not support routine testing for heritable thrombophilias as part of an evaluation for possible causes of stillbirth.
Introduction
Heritable thrombophilias are a heterogeneous group of conditions associated with an increased risk of thromboembolism. Typically they involve a decrease or abnormality in anticoagulant proteins or an increase in procoagulant factors. Acquired thrombophilias such as antiphospholipid syndrome have been associated with stillbirth, fetal growth restriction, and preeclampsia. In part, this is thought to be due to thrombosis in the uteroplacental circulation, leading to infarction, placental insufficiency, and fetal death. Indeed numerous studies found an association between heritable thrombophilias and stillbirth. However, other studies found no link between these conditions and stillbirth and the association remains controversial. Despite the lack of a clear association, many clinicians routinely screen for thrombophilias in women with adverse pregnancy outcomes including stillbirth.
Many studies have been limited by a small number of stillbirths, the inclusion of composite outcomes, limited detail regarding the stillbirths, and assessment of maternal but not fetal/placental thrombophilias. Assessment of fetal thrombophilias may be important because the placental tissue is genetically fetal. Thus, our objective was to assess the association between maternal and fetal/placental heritable thrombophilias and stillbirth in a population-based, case-control study in a geographically, racially, and ethnically diverse population.
Materials and Methods
The Stillbirth Collaborative Research Network (SCRN) of the Eunice Kennedy Shriver National Institute of Child Health and Human Development conducted a population-based case-control study of stillbirth from March 2006 through September 2008. Details of the study design were published. Participants were enrolled at the time of delivery and all gave written informed consent. The study was approved by the institutional review boards of the data coordinating center and each clinical site.
Attempts were made to enroll all deliveries with stillbirth (cases) and a contemporaneous representative sample of deliveries with live birth (controls) to women residing in SCRN catchment areas. These catchment areas included 59 tertiary care and community hospitals in portions of 5 states: Rhode Island, Massachusetts, Georgia, Texas, and Utah. The areas were defined by state and county boundaries, and the 59 hospitals within the catchment areas deliver a combined total of >80,000 infants per year. Some subgroups of live births were oversampled to ensure adequate numbers for stratified analyses.
Stillbirth was defined as Apgar scores of 0 at 1 and 5 minutes and no signs of life by direct observation at ≥20 weeks’ gestation. In addition, fetal deaths at 18 or 19 weeks without good dating were also included in the study to ensure that we did not miss losses that might prove to be >20 weeks’ gestation. Gestational age at delivery was determined by the best clinical estimate using multiple sources including assisted reproductive technology with documentation of the day of ovulation or embryo transfer (if available), first day of the last menstrual period, and results of obstetric sonograms. Deliveries resulting from the termination of a live fetus were excluded.
All cases and controls had a standardized maternal interview during the delivery hospitalization and detailed chart abstraction of prenatal office visits, antepartum hospitalizations, and the delivery hospitalization. Maternal race was self-reported. Cases and controls also had uniform placental pathology evaluation, and cases had comprehensive standardized fetal postmortem examination. Both were performed by a perinatal pathologist. Treating physicians occasionally tested for the presence of maternal thrombophilias after diagnosis of stillbirth as part of a clinically recommended evaluation for causes of stillbirth. However, testing was performed at the discretion of clinicians and was not done in each case. In addition, maternal blood for serum and DNA, fetal blood from the umbilical cord (when available), and placental tissue in all cases and controls and fetal tissue in cases were collected for research testing of thrombophilias. Maternal serum was stored at –80°C for 2-5 years prior to assay.
Stillbirths were classified using the Initial Causes of Fetal Death (INCODE) system. INCODE was developed to systematically assign causes of death using a priori definitions based on the best available evidence. Conditions were considered to be a probable cause of stillbirth if they had a high likelihood of directly causing the fetal death. Conditions that were not a direct cause of the stillbirth, but might be involved in a pathophysiologic sequence that led to the fetal death, were considered a possible cause of death. Finally, potentially important conditions that existed but did not meet criteria for probable or possible causes of death were recorded as present .
DNA was extracted from either frozen buffy coats or frozen placenta using a validated manufacturer’s protocol and Qiagen PureGen reagents (Qiagen Systems, Valencia, CA). Concentrations for the samples and an estimated purity (260-/280-nm ratio) were determined by ultraviolet–visible spectrophotometry (Nanodrop, Thermo Scientific, Wilmington, DE). Samples were analyzed by agarose gel electrophoresis to visualize sample integrity.
Samples were genotyped for 4 single nucleotide polymorphisms (factor V Leiden, rs6025; prothrombin G20210A, rs1799963; methylene tetrahydrofolate reductase [MTHFR] C677T, rs1801133; MTHFR A1298C, rs1801131) and 1 deletion/insertion polymorphism (plasminogen activator inhibitor [PAI] 4G/5G, rs1799768) using Taqman allelic discrimination chemistry (Life Technologies, Foster City, CA) with validated assays and the manufacturer’s protocols. Specifically, 6 ng of DNA was included in a 5-μL reaction with a final concentration of ×1 Taqman genotyping master mix and ×0.5 Taqman genotyping assay mix. The genotyping assay mix was specific to each tested polymorphism and contained both primers and allele-specific probes. After a 10-minute initial denaturation at 95°C, samples were amplified for 40 cycles of 95°C for 15 seconds and 60°C for 1 minute on a 9700 thermal cycler (Life Technologies). Genotypes were detected through endpoint analysis. Genotyping software (SDS V2.3; Life Technologies) was used to automatically determine sample genotypes through generation of cluster plots. Genotypes were verified and the polymorphisms were evaluated for deviation from Hardy-Weinberg equilibrium. All samples were genotyped in duplicate to ensure accurate genotyping. Laboratory personnel were blinded to the clinical status of the samples.
Results were restricted to singleton gestations. Data analyses were weighted for oversampling and other aspects of the study design as well as for differential consent using SUDAAN software, Version 10.0. Construction of the weights has been previously described. Odds ratios (OR) and 95% confidence intervals (CI) were calculated from univariate logistic regression models regressing stillbirth/live birth status on each marker–common homozygous, heterozygous, and uncommon homozygous (2 degrees of freedom [df]), and common homozygous vs other (1 df). For marker categories with no observations for the stillbirth group or the live birth group under study, an OR and P value could not be computed. However, in situations where it was informative, an upper bound on the P value was computed. This was accomplished by identifying 1 observation in the group with the smallest weight to be reclassified to have the unobserved category without making the resulting comparison more extreme (that is, more significant). In many circumstances, a suitable observation could not be found and an upper bound on the P value could not be reported. All tests were performed at a nominal significance level of α = 0.05. All single df tests were 2-sided. No correction was made for multiple comparisons.
Materials and Methods
The Stillbirth Collaborative Research Network (SCRN) of the Eunice Kennedy Shriver National Institute of Child Health and Human Development conducted a population-based case-control study of stillbirth from March 2006 through September 2008. Details of the study design were published. Participants were enrolled at the time of delivery and all gave written informed consent. The study was approved by the institutional review boards of the data coordinating center and each clinical site.
Attempts were made to enroll all deliveries with stillbirth (cases) and a contemporaneous representative sample of deliveries with live birth (controls) to women residing in SCRN catchment areas. These catchment areas included 59 tertiary care and community hospitals in portions of 5 states: Rhode Island, Massachusetts, Georgia, Texas, and Utah. The areas were defined by state and county boundaries, and the 59 hospitals within the catchment areas deliver a combined total of >80,000 infants per year. Some subgroups of live births were oversampled to ensure adequate numbers for stratified analyses.
Stillbirth was defined as Apgar scores of 0 at 1 and 5 minutes and no signs of life by direct observation at ≥20 weeks’ gestation. In addition, fetal deaths at 18 or 19 weeks without good dating were also included in the study to ensure that we did not miss losses that might prove to be >20 weeks’ gestation. Gestational age at delivery was determined by the best clinical estimate using multiple sources including assisted reproductive technology with documentation of the day of ovulation or embryo transfer (if available), first day of the last menstrual period, and results of obstetric sonograms. Deliveries resulting from the termination of a live fetus were excluded.
All cases and controls had a standardized maternal interview during the delivery hospitalization and detailed chart abstraction of prenatal office visits, antepartum hospitalizations, and the delivery hospitalization. Maternal race was self-reported. Cases and controls also had uniform placental pathology evaluation, and cases had comprehensive standardized fetal postmortem examination. Both were performed by a perinatal pathologist. Treating physicians occasionally tested for the presence of maternal thrombophilias after diagnosis of stillbirth as part of a clinically recommended evaluation for causes of stillbirth. However, testing was performed at the discretion of clinicians and was not done in each case. In addition, maternal blood for serum and DNA, fetal blood from the umbilical cord (when available), and placental tissue in all cases and controls and fetal tissue in cases were collected for research testing of thrombophilias. Maternal serum was stored at –80°C for 2-5 years prior to assay.
Stillbirths were classified using the Initial Causes of Fetal Death (INCODE) system. INCODE was developed to systematically assign causes of death using a priori definitions based on the best available evidence. Conditions were considered to be a probable cause of stillbirth if they had a high likelihood of directly causing the fetal death. Conditions that were not a direct cause of the stillbirth, but might be involved in a pathophysiologic sequence that led to the fetal death, were considered a possible cause of death. Finally, potentially important conditions that existed but did not meet criteria for probable or possible causes of death were recorded as present .
DNA was extracted from either frozen buffy coats or frozen placenta using a validated manufacturer’s protocol and Qiagen PureGen reagents (Qiagen Systems, Valencia, CA). Concentrations for the samples and an estimated purity (260-/280-nm ratio) were determined by ultraviolet–visible spectrophotometry (Nanodrop, Thermo Scientific, Wilmington, DE). Samples were analyzed by agarose gel electrophoresis to visualize sample integrity.
Samples were genotyped for 4 single nucleotide polymorphisms (factor V Leiden, rs6025; prothrombin G20210A, rs1799963; methylene tetrahydrofolate reductase [MTHFR] C677T, rs1801133; MTHFR A1298C, rs1801131) and 1 deletion/insertion polymorphism (plasminogen activator inhibitor [PAI] 4G/5G, rs1799768) using Taqman allelic discrimination chemistry (Life Technologies, Foster City, CA) with validated assays and the manufacturer’s protocols. Specifically, 6 ng of DNA was included in a 5-μL reaction with a final concentration of ×1 Taqman genotyping master mix and ×0.5 Taqman genotyping assay mix. The genotyping assay mix was specific to each tested polymorphism and contained both primers and allele-specific probes. After a 10-minute initial denaturation at 95°C, samples were amplified for 40 cycles of 95°C for 15 seconds and 60°C for 1 minute on a 9700 thermal cycler (Life Technologies). Genotypes were detected through endpoint analysis. Genotyping software (SDS V2.3; Life Technologies) was used to automatically determine sample genotypes through generation of cluster plots. Genotypes were verified and the polymorphisms were evaluated for deviation from Hardy-Weinberg equilibrium. All samples were genotyped in duplicate to ensure accurate genotyping. Laboratory personnel were blinded to the clinical status of the samples.
Results were restricted to singleton gestations. Data analyses were weighted for oversampling and other aspects of the study design as well as for differential consent using SUDAAN software, Version 10.0. Construction of the weights has been previously described. Odds ratios (OR) and 95% confidence intervals (CI) were calculated from univariate logistic regression models regressing stillbirth/live birth status on each marker–common homozygous, heterozygous, and uncommon homozygous (2 degrees of freedom [df]), and common homozygous vs other (1 df). For marker categories with no observations for the stillbirth group or the live birth group under study, an OR and P value could not be computed. However, in situations where it was informative, an upper bound on the P value was computed. This was accomplished by identifying 1 observation in the group with the smallest weight to be reclassified to have the unobserved category without making the resulting comparison more extreme (that is, more significant). In many circumstances, a suitable observation could not be found and an upper bound on the P value could not be reported. All tests were performed at a nominal significance level of α = 0.05. All single df tests were 2-sided. No correction was made for multiple comparisons.
Results
The flow of participants into the SCRN study and this analysis is shown in the Figure . Maternal thrombophilia testing was successful for ≥1 marker in 488 singleton cases and 1342 singleton controls. Fetal thrombophilia testing was successful for 405 cases and 990 controls. We combined the results for thrombophilia testing in DNA obtained from either the fetus or placenta and considered it to be fetal.
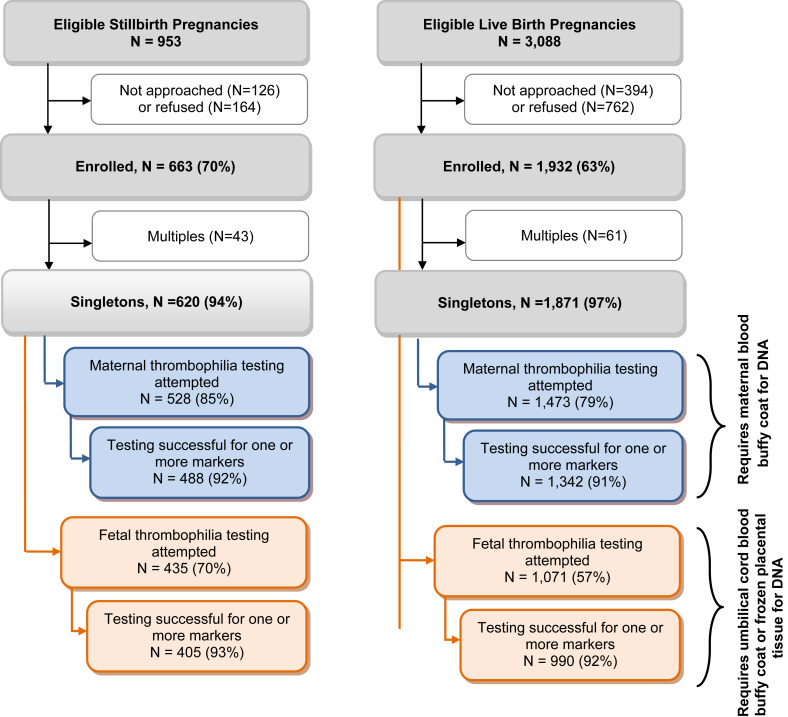
Characteristics of the cohort have previously been described in detail. Characteristics of the women included in this analysis with and without successful thrombophilia testing on the maternal and/or fetal samples are shown in Supplemental Tables 1 and 2 , respectively. Slight differences among groups with regard to demographics are noted in the tables.
Table 1 shows the results of maternal thrombophilia testing in all stillbirths compared to all live births, and all stillbirths compared to term live births. Overall, there was a significant association between factor V Leiden genotype and stillbirth compared with live births ( P = .0004). There was a significant increased odds of stillbirth for the homozygous mutation (2/488; 0.4% vs 1/1380; 0.0046% OR, 87.44; 95% CI, 7.88–970.92). Of the 3 women homozygous for the factor V Leiden mutation, 2 had term antepartum stillbirths and 1 had a live birth delivered at 22 weeks’ gestation. There was a similar association that was not significant for the heterozygous mutation (5.2% vs 3.3%, OR, 1.61; 95% CI, 0.90–2.90). The groups were similar for other thrombophilias including the prothrombin G20210A, PAI-1 4G/5G, MTHFR A1298C, and MTHFR C677T mutations. Results were similar for comparison of stillbirths with term live births. When stratified by race/ethnicity, factor V Leiden remained associated with stillbirth in non-Hispanic white women, but not non-Hispanic black or Hispanic women (data not shown). All 3 of the factor V Leiden homozygous mutations were in non-Hispanic white women.
| Characteristic–weighted % b | Stillbirth N = 488 N w = 487 | Live birth N = 1342 N w = 1037 | Odds ratio (CI) | P value | Term live birth N = 1026 N w = 933 | Odds ratio (CI) | P value |
|---|---|---|---|---|---|---|---|
| Factor V Leiden G508A (rs6025) | |||||||
| GG | 94.4 | 96.7 | reference | .0004 | 96.8 | reference | .0237 d |
| AG | 5.2 | 3.3 | 1.61 (0.90–2.90) | 3.2 | 1.64 (0.89–3.03) | ||
| AA | 0.4 | 0.0 c | 87.44 (7.88–970.92) | 0 | — | ||
| Common homozygous vs other | |||||||
| GG | 94.4 | 96.7 | reference | .0611 | 96.8 | reference | .0627 |
| Other | 5.6 | 3.3 | 1.73 (0.97–3.08) | 3.2 | 1.77 (0.97–3.22) | ||
| FII prothrombin G20210 (rs1799963) | |||||||
| AA | 0.2 | 0.1 | 1.72 (0.11–27.72) | .9232 | 0.1 | 1.54 (0.10–24.81) | .9467 |
| AG | 1.6 | 1.6 | 1.05 (0.44–2.54) | 1.7 | 0.95 (0.39–2.29) | ||
| GG | 98.2 | 98.3 | reference | 98.1 | reference | ||
| Common homozygous vs other | |||||||
| GG | 98.2 | 98.3 | reference | .8295 | 98.1 | reference | .9750 |
| Other | 1.8 | 1.7 | 1.10 (0.47–2.54) | 1.9 | 0.99 (0.43–2.29) | ||
| PAI-1 4G/5G in/del (rs1799768) | |||||||
| 5G/5G (G/G) | 39.1 | 34.7 | reference | .0611 | 34.4 | reference | .1083 |
| 4G/5G (−/G) | 41.7 | 40.3 | 0.92 (0.70–1.20) | 41.6 | 0.88 (0.67–1.16) | ||
| 4G/4G (−/−) | 19.1 | 25.0 | 0.68 (0.49–0.94) | 24.0 | 0.70 (0.50–0.98) | ||
| Common homozygous vs other | |||||||
| 5G/5G (G/G) | 39.1 | 34.7 | reference | .1305 | 34.4 | reference | .1124 |
| Other | 60.9 | 65.3 | 0.83 (0.65–1.06) | 65.6 | 0.82 (0.63–1.05) | ||
| MTHFR A1298C (rs1801131) | |||||||
| CC | 6.0 | 6.6 | 0.91 (0.56–1.48) | .8183 | 6.7 | 0.90 (0.55–1.48) | .8374 |
| AC | 32.5 | 31.1 | 1.06 (0.82–1.36) | 31.3 | 1.05 (0.81–1.36) | ||
| AA | 61.5 | 62.3 | reference | 62.0 | reference | ||
| Common homozygous vs other | |||||||
| AA | 61.5 | 62.3 | reference | .8033 | 62.0 | reference | .8699 |
| Other | 38.5 | 37.7 | 1.03 (0.81–1.31) | 38.0 | 1.02 (0.80–1.31) | ||
| MTHFR C677T (rs1801133) | |||||||
| CC | 46.8 | 41.5 | reference | .1087 | 41.1 | reference | .1367 |
| CT | 40.3 | 41.9 | 0.85 (0.66–1.10) | 43.2 | 0.82 (0.63–1.07) | ||
| TT | 12.9 | 16.6 | 0.69 (0.48–0.99) | 15.7 | 0.72 (0.50–1.04) | ||
| Common homozygous vs other | |||||||
| CC | 46.8 | 41.5 | reference | .0759 | 41.1 | reference | .0622 |
| Other | 53.2 | 58.5 | 0.81 (0.63–1.02) | 58.9 | 0.79 (0.62–1.01) |
a Stillbirth Collaborative Research Network (SCRN) defined stillbirth as Apgar scores of 0/0 at 1 and 5 min with no signs of life by direct observation. Results are presented here for pregnancies. Pregnancy is defined as SCRN case if any stillbirths delivered and SCRN control if all live births delivered. SCRN case status is labeled “stillbirth” or “live birth”
b Results shown (percentages, odds ratios, and P values) are weighted for study design and differential consent based on characteristics recorded on all eligible pregnancies screened for study. Unweighted (N) and weighted (N w ) sample sizes are also provided. Sample sizes vary slightly by characteristic included in table
c Percent denoted “0.0” indicates at least 1 observation in category (rounded to 0.0), whereas percent denoted “0” indicates no observation in category. For thrombophilia marker categories with no observations for stillbirth group or live birth group, odds ratio and P value cannot be computed
d However, in situations where it is informative, upper bound on P value has been computed and is reported.
Comparisons of fetal thrombophilias in stillbirths and live births as well as stillbirths and term live births are shown in Table 2 . There were decreased odds of stillbirth for fetal 4G/4G PAI-1 polymorphism compared to term live births (OR, 0.63; 95% CI, 0.43–0.91) and with heterozygous (CT) or homozygous (TT) mutation MTHFR C677T compared to all live births (OR, 0.76; 95% CI, 0.59–0.98). All other thrombophilias were similar among groups. There were no differences in fetal thrombophilias between stillbirths and live births or term live births for any thrombophilia when stratified by race/ethnicity (data not shown).
| Characteristic–weighted % b | Stillbirth N = 405 N w = 395 | Live birth N = 990 N w = 827 | Odds ratio (CI) | P value | Term live births N = 841 N w = 776 | Odds ratio (CI) | P value |
|---|---|---|---|---|---|---|---|
| Factor V Leiden G508A (rs6025) | |||||||
| GG | 96.8 | 97.2 | reference | — | 97.2 | reference | — |
| AG | 3.2 | 2.6 | 1.25 (0.58–2.67) | 2.7 | 1.23 (0.57–2.65) | ||
| AA | 0 | 0.2 | — | 0.2 | — | ||
| Common homozygous vs other | |||||||
| GG | 96.8 | 97.2 | reference | .6786 | 97.2 | reference | .7187 |
| Other | 3.2 | 2.8 | 1.17 (0.55–2.49) | 2.8 | 1.15 (0.54–2.46) | ||
| FII prothrombin G20210 (rs1799963) | |||||||
| AA | 0 | 0 | — | — | 0 | — | — |
| AG | 1.9 | 1.4 | — | 1.4 | — | ||
| GG | 98.1 | 98.6 | — | 98.6 | — | ||
| Common homozygous vs other | |||||||
| GG | 98.1 | 98.6 | reference | .5432 | 98.6 | reference | .5010 |
| Other | 1.9 | 1.4 | 1.35 (0.51–3.57) | 1.4 | 1.41 (0.52–3.81) | ||
| PAI-1 4G/5G in/del (rs1799768) | |||||||
| 5G/5G (G/G) | 41.4 | 35.5 | reference | .0558 | 35.2 | reference | .0472 |
| 4G/5G (−/G) | 41.1 | 41.0 | 0.86 (0.64–1.15) | 41.0 | 0.85 (0.63–1.14) | ||
| 4G/4G (−/−) | 17.5 | 23.5 | 0.64 (0.44–0.92) | 23.7 | 0.63 (0.43–0.91) | ||
| Common homozygous vs other | |||||||
| 5G/5G (G/G) | 41.4 | 35.5 | reference | .0677 | 35.2 | reference | .0589 |
| Other | 58.6 | 64.5 | 0.78 (0.60–1.02) | 64.8 | 0.77 (0.59–1.01) | ||
| MTHFR A1298C (rs1801131) | |||||||
| CC | 6.5 | 8.0 | 0.82 (0.50–1.34) | .6425 | 8.2 | 0.79 (0.48–1.30) | .5952 |
| AC | 33.4 | 31.9 | 1.05 (0.80–1.38) | 32.1 | 1.03 (0.78–1.36) | ||
| AA | 60.0 | 60.1 | reference | 59.6 | reference | ||
| Common homozygous vs other | |||||||
| AA | 60.0 | 60.1 | reference | .9853 | 59.6 | reference | .9039 |
| Other | 40.0 | 39.9 | 1.00 (0.78–1.29) | 40.4 | 0.98 (0.76–1.28) | ||
| MTHFR C677T (rs1801133) | |||||||
| CC | 51.9 | 45.1 | reference | .0718 | 46.0 | reference | .1263 |
| CT | 35.0 | 37.9 | 0.80 (0.61–1.07) | 37.2 | 0.83 (0.63–1.11) | ||
| TT | 13.0 | 17.0 | 0.66 (0.45–0.97) | 16.8 | 0.69 (0.47–1.01) | ||
| Common homozygous vs other | |||||||
| CC | 51.9 | 45.1 | reference | .0367 | 46.0 | reference | .0723 |
| Other | 48.1 | 54.9 | 0.76 (0.59–0.98) | 54.0 | 0.79 (0.61–1.02) |
a Stillbirth Collaborative Research Network (SCRN) defined stillbirth as Apgar scores of 0/0 at 1 and 5 min with no signs of life by direct observation. Results are presented here for pregnancies. Pregnancy is defined as SCRN case if any stillbirths delivered and SCRN control if all live births delivered. SCRN case status is labeled “stillbirth” or “live birth”
b Results shown (percentages, odds ratios, and P values) are weighted for study design and differential consent based on characteristics recorded on all eligible pregnancies screened for study. Unweighted (N) and weighted (N w ) sample sizes are also provided. Sample sizes vary slightly by characteristic included in table. Percent denoted “0” indicates no observation in category. For thrombophilia marker categories with no observations for stillbirth group or live birth group, odds ratio and P value cannot be computed.
Subanalyses on maternal and fetal samples for stillbirths vs live births excluding fetal anomalies and intrapartum stillbirths are shown in Table 3 . In this subset of cases and controls, maternal factor V Leiden genotype was associated with stillbirth ( P = .0001). Homozygous factor V Leiden mutation was significantly associated with stillbirth (0.5% vs 0.0048%; OR, 118.96; 95% CI, 10.71–1321.88). Heterozygous mutation did not reach statistical significance (5.8% vs 3.3% heterozygous mutation; OR, 1.82; 95% CI, 0.97–3.39) No other maternal thrombophilias or any fetal thrombophilias were associated with stillbirth in these subanalyses.



