What are the different types of microarray?
There are 2 major microarray platforms used in prenatal diagnosis: single-nucleotide polymorphism (SNP) arrays and comparative genomic hybridization (CGH) arrays. With SNP and CGH arrays, DNA from a fetal sample, such as CVS or amniocentesis, is hybridized to an array platform consisting of DNA probes on a solid surface, such as a microscope slide or a silicon chip.
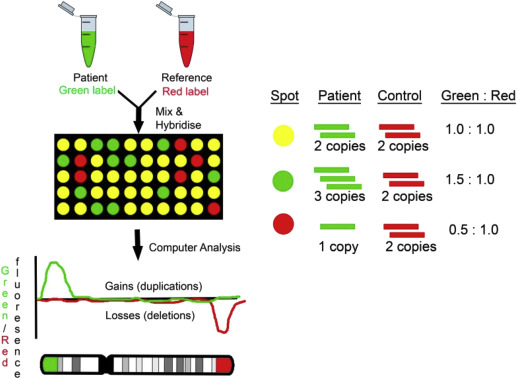
CGH compares the fetal DNA sample with a normal reference DNA sample ( Figure 1 ). The test DNA and the reference DNA samples are labeled with 2 different-colored fluorescent dyes, then combined and hybridized to an array platform. The relative intensities of the different colors are compared with bioinformatics tools. Cases with duplications will have a greater hybridization signal, whereas cases with deletions will have a lower hybridization signal compared to the reference sample.
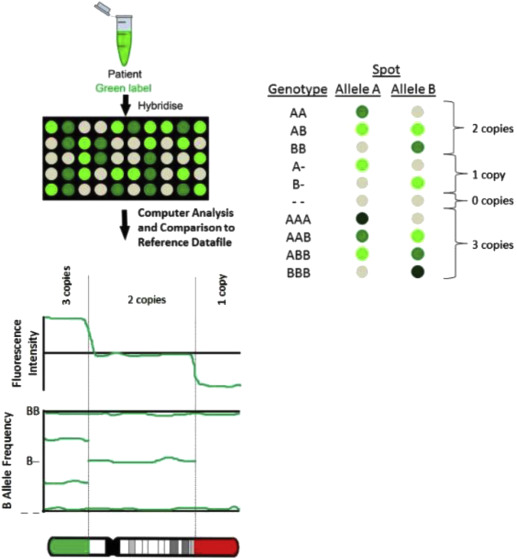
A SNP is a variation at a single position in a DNA sequence among individuals. With SNP arrays, only the DNA test sample is hybridized to the array platform ( Figure 2 ). SNP arrays detect CNVs by measuring probe signal intensities as used in the CGH approach. Although CGH arrays are only able to detect CNVs, SNP arrays also can detect triploidy and regions on the 2 homologous chromosomes that are identical to each other, as occurs with uniparental disomy (UPD) and consanguinity. With UPD, both copies of a chromosome are inherited from the same parent instead of 1 from each parent. UPD has been associated with genetic disorders such as Prader-Willi syndrome, which can occur when both copies of chromosomes 15 are inherited maternally. SNP arrays also can detect some cases of maternal cell contamination and mosaicism.
Arrays may include probes that cover the whole genome, or may be targeted with concentrated coverage in known disease-causing regions of the genome and more limited coverage of the rest of the genome. An advantage of targeted arrays is that they decrease the chance of identifying a variant of uncertain significance (see section “What are the risks of using CMA? What are variants of uncertain significance and how should they be managed?”). In general, arrays used for prenatal diagnosis have lower resolution than those used for postnatal testing for this reason.
What can CMA detect? How does it differ from a karyotype?
A standard karyotype can detect aneuploidies (abnormalities in chromosome number), relatively large structural abnormalities such as deletions or duplications that are microscopically visible down to a resolution of approximately 5−10 Mb, and balanced or unbalanced translocations and inversions. CMA has a greater resolution than conventional karyotyping, allowing for the detection of much smaller, submicroscopic deletions, and duplications typically down to a 50- to 100-kb level. CMA also can detect some copy number changes near the chromosomal breakpoint sites in rearrangements that appear to be balanced on a conventional karyotype. The types of abnormalities that can be detected with CGH and SNP arrays and conventional karyotype are summarized in Table 1 .
| Technique | Aneuploidy | Balanced translocations and inversions | Unbalanced translocations | Triploidy | AOH/consanguinity | CNVs |
|---|---|---|---|---|---|---|
| Conventional karyotype | + | + | + | + | − | − |
| CGH array | + | − | + | − | − | + |
| SNP array | + | − | + | + | + | + |
Several large-scale studies have compared the prenatal use of chromosomal microarray to conventional karyotyping. A 2012 multicenter trial sponsored by the National Institute of Child Health and Human Development (NICHD) demonstrated that CMA is most beneficial in fetuses with abnormal ultrasound findings. In pregnancies with a fetal structural abnormality identified by ultrasound and a normal karyotype, CMA revealed clinically relevant deletions or duplications in 6.0% of cases. In pregnancies with a structurally normal fetus by ultrasound and a normal karyotype in which CVS or amniocentesis was performed secondary to advanced maternal age or a positive aneuploidy screen, CMA revealed clinically relevant findings in 1.7% of cases. A systematic review of 4 large-scale studies, including the NICHD trial, reported clinically significant deletions or duplications in 6.5% (201/3090) of cases with abnormal ultrasound findings, 1.0% (50/5108) of cases with advanced maternal age, and 1.1% (44/4164) of cases in which CVS or amniocentesis was performed as the result of another reason, including parental anxiety, an abnormal serum screening result, or history of a chromosome abnormality. All of these cases had normal karyotypes.
Another advantage of CMA is that this technique does not require dividing cells, in contrast to conventional karyotyping, which requires cell culture. This difference can allow a quicker turnaround time. In addition, CMA can be performed on macerated tissue obtained from stillbirth specimens that may not grow in tissue culture; therefore, CMA may be more likely to provide a result as long as sufficient good-quality DNA can be obtained. A population-based study of 532 stillbirth cases by the Stillbirth Collaborative Research Network reported that microarray analysis yielded results more often than standard karyotype analysis (87.4% vs 70.5%, P < .0001) and provided increased detection of genetic abnormalities (8.3% vs 5.8%, P = .0067). We recommend that CMA be offered when genetic analysis is performed in cases with fetal structural anomalies and/or stillbirth and replaces the need for fetal karyotype in these cases (GRADE 1A) .
What can CMA detect? How does it differ from a karyotype?
A standard karyotype can detect aneuploidies (abnormalities in chromosome number), relatively large structural abnormalities such as deletions or duplications that are microscopically visible down to a resolution of approximately 5−10 Mb, and balanced or unbalanced translocations and inversions. CMA has a greater resolution than conventional karyotyping, allowing for the detection of much smaller, submicroscopic deletions, and duplications typically down to a 50- to 100-kb level. CMA also can detect some copy number changes near the chromosomal breakpoint sites in rearrangements that appear to be balanced on a conventional karyotype. The types of abnormalities that can be detected with CGH and SNP arrays and conventional karyotype are summarized in Table 1 .
| Technique | Aneuploidy | Balanced translocations and inversions | Unbalanced translocations | Triploidy | AOH/consanguinity | CNVs |
|---|---|---|---|---|---|---|
| Conventional karyotype | + | + | + | + | − | − |
| CGH array | + | − | + | − | − | + |
| SNP array | + | − | + | + | + | + |
Several large-scale studies have compared the prenatal use of chromosomal microarray to conventional karyotyping. A 2012 multicenter trial sponsored by the National Institute of Child Health and Human Development (NICHD) demonstrated that CMA is most beneficial in fetuses with abnormal ultrasound findings. In pregnancies with a fetal structural abnormality identified by ultrasound and a normal karyotype, CMA revealed clinically relevant deletions or duplications in 6.0% of cases. In pregnancies with a structurally normal fetus by ultrasound and a normal karyotype in which CVS or amniocentesis was performed secondary to advanced maternal age or a positive aneuploidy screen, CMA revealed clinically relevant findings in 1.7% of cases. A systematic review of 4 large-scale studies, including the NICHD trial, reported clinically significant deletions or duplications in 6.5% (201/3090) of cases with abnormal ultrasound findings, 1.0% (50/5108) of cases with advanced maternal age, and 1.1% (44/4164) of cases in which CVS or amniocentesis was performed as the result of another reason, including parental anxiety, an abnormal serum screening result, or history of a chromosome abnormality. All of these cases had normal karyotypes.
Another advantage of CMA is that this technique does not require dividing cells, in contrast to conventional karyotyping, which requires cell culture. This difference can allow a quicker turnaround time. In addition, CMA can be performed on macerated tissue obtained from stillbirth specimens that may not grow in tissue culture; therefore, CMA may be more likely to provide a result as long as sufficient good-quality DNA can be obtained. A population-based study of 532 stillbirth cases by the Stillbirth Collaborative Research Network reported that microarray analysis yielded results more often than standard karyotype analysis (87.4% vs 70.5%, P < .0001) and provided increased detection of genetic abnormalities (8.3% vs 5.8%, P = .0067). We recommend that CMA be offered when genetic analysis is performed in cases with fetal structural anomalies and/or stillbirth and replaces the need for fetal karyotype in these cases (GRADE 1A) .
What are the limitations of CMA?
Because CMA looks for genomic imbalance, this technique is not able to detect totally balanced chromosomal rearrangements, such as translocations or inversions. The large majority of balanced rearrangements, however, result in a normal outcome. In addition, CMA does not provide information about the chromosomal mechanism of a genetic imbalance. For example, if there is a gain of an entire chromosome 13, CMA cannot distinguish between trisomy 13 and an unbalanced Robertsonian translocation, which has relevance for recurrence risk counseling. Therefore, a karyotype should be performed in such cases to rule out a translocation that may have been inherited. Low-level mosaicism may not be detected by CMA, and some arrays do not detect triploidy. SNP arrays, however, generally are able to detect lower levels of mosaicism, as well as triploidy. CMA will not detect all CNVs, such as those that are in regions not represented on the array platform and very small CNVs that are below the level of detection. In some cases, a postnatal CMA may identify a CNV that was not identified prenatally because of the greater resolution of postnatal arrays. In addition, CMA will not detect point mutations within single genes, including those that cause disorders such as sickle-cell anemia, cystic fibrosis, and many of the skeletal dysplasias.
What are the risks of using CMA? What are variants of uncertain significance and how should they be managed?
A disadvantage of CMA is the inability to precisely interpret the clinical significance of a previously unreported CNV or to accurately predict the phenotype of some CNVs that are associated with variable outcomes. CNVs are characterized as benign, clinically significant (ie, pathogenic), and as a variant of uncertain significance (VUS). The overall prevalence of VUS is approximately 1−2%. The NICHD-sponsored multicenter trial reported a 0.9% prevalence of pathogenic CNVs and a 1.5% prevalence of VUS in cases with normal karyotypes. A qualitative study of 12 couples who received a pathogenic (n = 6) or uncertain microarray result (n = 6) reported that participants whose fetuses carried a VUS experienced heightened anxiety and frustration at the limited scope of information available to them to understand and plan for the future health and development of their child and make decisions about continuing the pregnancy. Some of these couples felt “even more uninformed after microarray.” Fortunately, additional information on the classification of CNVs is accumulating rapidly, which should lead to a decrease in the incidence of reported VUS over time.
We recommend that patients in whom a fetal VUS is detected receive counseling from experts who have access to databases that provide updated information concerning genotype-phenotype correlations (Best Practice) . Patients should be educated regarding the significance of the finding, including the potential range of outcomes, and should be provided with resources and support. Further testing should be offered if indicated. One of the initial steps in the evaluation is to determine whether either parent has the same CNV as was detected in the fetus. Although de novo CNVs are more likely to be pathogenic, an abnormal fetal outcome cannot always be excluded even if a parent with the same CNV as a fetus is normal, as some have a variable outcome. When interpreting VUS, it may be helpful to evaluate the specific genes that are contained in the deleted or duplicated regions. In general, small duplications are less likely to be clinically significant than small deletions.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


