Preimplantation genetic testing for aneuploidy
The evolution of aneuploidy screening techniques has provided more reliable and faster results about genetic status of the embryos. It has been estimated that approximately 100,000 PGD cycles have been performed worldwide over the past 23 years and nearly 80% of these cycles have been PGT-A [7]. Currently, PGT-A constitutes the majority of PGD cycles performed globally. PGT-A is also controversial in terms of the effectiveness of comprehensive techniques used in different patient groups and using different cell types for biopsy.
24.2 Biopsy
24.2.1 The Source of Sample
There are currently three sources of cellular materials that can be used for PGT-A. Polar body (PB) is usually retrieved from oocytes and zygotes. This approach restricts detection of chromosome abnormalities to the female. Blastomere biopsy involves the removal of a single cell from a day 3 stage embryo. This approach allows identification of both maternal and paternal contributions. Embryo transfer can be performed 2 days later, on day 5 of development. However, this approach cannot detect mosaicism and may damage the embryo viability.
Improvements in embryo culture conditions as well as in vitrification systems have paved the way for the current trend of trophectoderm (TE) biopsies. TE biopsy is performed at the blastocyst stage, on day 5, day 6, or sometimes day 7. At this stage, embryos have undergone their first cellular differentiation, resulting in two cell lineages: the inner cell mass (ICM) and TE cells. Biopsy of TE cells does not adversely affect embryo development. With TE biopsy, multiple cells can be sampled from each embryo, which can improve accuracy in the genetics laboratory. Current most PGT-A research focuses on genome-wide aneuploidy screening of biopsies from the TE, which may better represent the ultimate genetic constitution of the embryo. More recently, new types of sample, like blastocentesis and analysis of spent embryo culture medium, have also been proposed. However, further results are needed to ensure reliability of the results before widespread clinical application [8, 9].
24.2.2 Sampling Method
For each biopsy approach, the zona pellucida (ZP) must be perforated. This can be accomplished by several methods, including mechanically cutting through the ZP with a micropipette, chemically dissolving the ZP with a weak acid solution (acid Tyrode’s), or laser via the optical system of a microscope. The laser approach is most commonly used because it is faster, safer, and more reproducible among technicians.
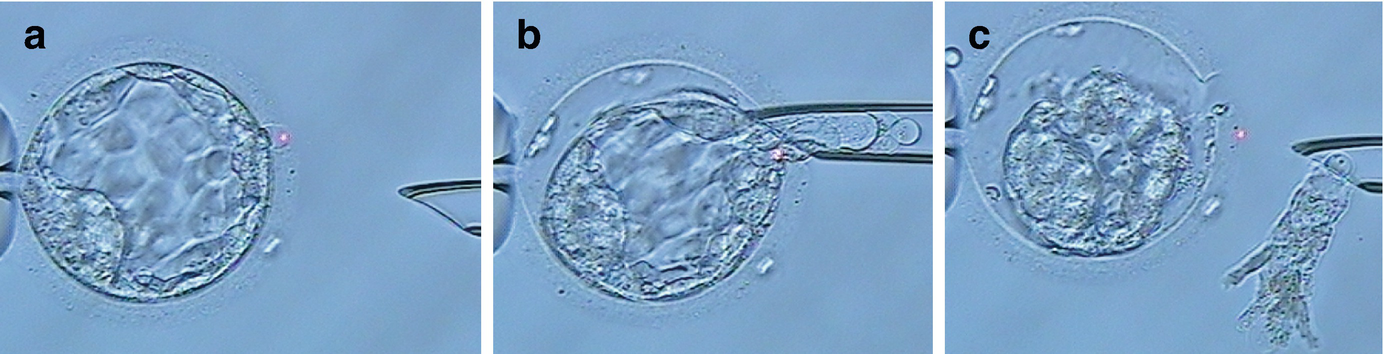
Blastocyst biopsy is performed by simultaneous zona pellucida opening and trophectoderm biopsy. (a) Laser-assisted hatching on day 5 blastocyst, (b) 5–10 trophectoderm cells were sucked into the biopsy pipette and then targeted with few laser pulses, (c) trophectoderm cells are dissected from the blastocyst by gentle suction during the laser firing
24.3 Diagnostic Technologies
24.3.1 FISH
FISH with chromosome-specific DNA probes can be applied to polar body, blastomere, or TE cell that had been fixed on slides and gives detectable signals in interphase nuclei as well as on metaphase chromosomes. However, The FISH technique is limited by having relatively few discrete colors available, and typically up to five chromosomes can be tested at the same time. The same nucleus can be retested (reprobed), and in this way, 12 chromosomes have been tested in clinical practice. In addition, FISH requires high technical skills and scoring FISH signals in a single nucleus is inherently subjective and prone to errors.
24.3.2 aCGH
aCGH is the first technology to be widely available for reliable, accurate, and relatively fast 24-chromosome copy number analysis. In this method, the biopsied cells are amplified using a WGA technique. Amplified sample DNA and control DNA are differentially labeled with fluorophores and then competitively hybridized on the array platform at the same time. Chromosome loss or gain is revealed by the color of each spot after hybridization. This is because the technique involves the competitive hybridization of differentially labeled test and reference euploid DNA samples. Fluorescence intensity is detected using a laser scanner and data processing software, which can analyze whole chromosome aneuploidy and sub-chromosomal structural imbalances. aCGH is now used extensively around the world despite the relatively high cost of testing multiple samples [12].
24.3.3 SNP Array
Single nucleotide polymorphism (SNP) is a DNA sequence variant in which, at a particular position or locus, one of two or more nucleotides may be present on different chromosomes within a population. SNP arrays involves tracking the inheritance of polymorphisms from the parents to their embryos. Each of the parental chromosomes has a unique combination of SNP alleles providing a means of confirming whether a particular chromosome is present or absent from a sample. The process of SNP array involves WGA, fluorescent labeling of the amplified embryo DNA, and ascertainment of SNP genotypes using a microarray. The results from embryos are compared with data obtained from the mother and father and ploidy status can be inferred and assigned to each embryo. A significant advantage of SNP array is that the simultaneous analysis of thousands of polymorphisms scattered throughout the genome produces a unique DNA fingerprint for each embryo tested. The DNA fingerprint allows parental origin to be confirmed, reducing the risk that a laboratory error could lead to embryos being transferred to the wrong patient. The main drawbacks of SNP array are that the test is more expensive and takes longer to perform than aCGH or qPCR [13].
24.3.4 qPCR
qPCR technique is a robust, rapid, accurate, and cost-effective comprehensive chromosome screening method. Briefly, a preamplification step involving the multiplex amplification of 96 loci is performed with the use of TaqMan copy number assays. The preamplified products are then quantified using RT-qPCR in a 384-well plate, and whole chromosome aneuploidies are determined. The whole procedure lasts about 4 h and can also be combined with mutation detection. In this approach, PCR is performed directly on the sample, without WGA, which demonstrates the biggest advantage of this technique. However, the main drawbacks of this system are the facts that it is unable to detect segmental abnormalities and has yet to be validated for the detection of mosaicism [7, 14].
24.3.5 NGS
Most recently, next-generation sequencing (NGS) has been applied to PGT-A as a potentially more efficient and affordable technique. NGS is based upon the ability to massively parallel sequence small DNA fragments until the required depth of coverage is attained. Succeeding WGA, a barcoding step follows to allow the identification of embryo-specific sequences after which the amplified product is broken down into small sequence-ready fragments. Those fragments are then subjected to massively parallel sequencing with low coverage for the purpose of aneuploidy screening. The number of reads per chromosome (“binning”) is proportional to the copy number of each chromosome and serves as a basis for aneuploidy calls [15]. The most important benefit of NGS is that it has the power of simultaneous assessment of aneuploidy, translocations, single-gene disorders, small copy number variations, and low-level mosaicism (<25%) from the same biopsy sample using the same platform technology [16]. Another benefit is, with NGS, a large number of samples can be simultaneously tested which results in reducing the cost and workload.
24.4 Clinical Outcome of PGT-A
Currently, multiple reports including meta-analysis and systematic reviews have demonstrated an improvement on implantation, clinical pregnancy, ongoing pregnancy, and live birth rates while reducing miscarriage rates and multiple pregnancy rates through the use of PGT-A [17, 18]. A randomized controlled trial (RCT) comparing blastocyst-stage single embryo fresh transfer with and without aCGH in good prognosis patients showed a significantly higher clinical pregnancy rate in the PGT-A group (70.9% vs. 45.8%). PGT-A group yielded a lower miscarriage rate than those without PGT-A [19]. A combination of the findings of 19 articles were summarized including 3 RCTs and 16 observational studies which revealed that in both young and advanced maternal age (AMA) patient populations, PGT-A results in a higher delivery rate per embryo transferred compared to the traditional method of morphology-based selection of embryos [20]. However, the data on patients of AMA, recurrent miscarriage, and implantation failure were from matched cohort studies, limiting their validity to make decisive conclusions. Similarly, in a meta-analysis where four RCTs and seven cohort studies were assessed for the effectiveness of PGT-A over traditional morphological methods, according to that, the transfer of euploid embryos can improve the implantation rate [21].
24.5 The Implementation Dilemma on Diminished Ovarian Reserve Patients
Despite the large and rapidly growing weight of evidence in support of PGT-A, some clinicians continue to have reservations about the clinical utility of this technique, particularly on diminished ovarian reserve (DOR) patients [22].
So why is PGT-A so controversial in DOR patients? One of the main reasons is that DOR patients have a reduced potential to produce an adequate number of oocytes and hence embryos, which may not allow PGT-A to be applied. Actually, many DOR patients who undergo IVF-PGT-A do not reach embryo transfer owing to the culmination of low embryo numbers and high aneuploidy rate in tested blastocyst-stage embryos. Shahine et al. found that the risk of not having a euploid blastocyst available for transfer in an IVF-PGT-A cycle was 13% in the normal ovarian reserve group and 25% in the DOR group. After including the eight patients with DOR who did not even make it to retrieval, the risk of no transfer for DOR patients was at least 37% [23]. It is important to note that the average number of oocytes retrieved was eight, and the mean number of blastocysts biopsied was 3.6 in this study, which still is a reasonable number to proceed with IVF-PGT-A. Some patients with severe DOR will have even fewer oocytes, blastocysts, and higher likelihood of no blastocyst formation. So when counseling DOR patients about the success rates with IVF-PGT-A, it is important to quote not only the success rates per transfer but also the likelihood of having an euploid blastocyst available for transfer [24]. There are two studies that provided data comparing PGT-A with standard morphology assessment among anticipated poor responders. The authors reported that although more poor responders in the PGT-A arm had no transfer performed because of no euploid embryos being available, the PGT-A arm had a higher delivery rate per randomized patient (36 versus 21.9%, P < 0.05). This improvement was because of a significantly higher pregnancy rate per transfer (52.9 versus 24.2%, P < 0.001) and a significant reduction in miscarriage (2.7 versus 39%, P < 0.001) in the PGT-A arm [25]. Indeed, many patients randomized to PGT-A in such a study will have no embryos available to transfer if the only clinically usable embryos are found to be aneuploid. The true benefit in such patients may be the result of avoiding futile transfers and expeditiously moving into either another stimulation cycle or egg/embryo donation. Thus, time to pregnancy may be a better metric with miscarriage rate being a useful secondary measure [26].
Another reason for the controversy is the accuracy of the PGT-A’s diagnostic results. The importance of avoiding a false diagnosis of euploidy and eliminating the chance of an aneuploid ongoing gestation is obvious. However, given the small number of embryos available for transfer in DOR patients, the avoidance of a false diagnosis of aneuploidy is very important in this patient population. Indeed, discarding an embryo because of incorrectly labeling it as abnormal may eliminate a patient’s only chance at transfer. The chance of misdiagnosis results from both technical and biological limitations. Technical error can arise with any screening platform and occurs because of DNA contamination, allele dropout, human error, failed amplification, or a variety of other causes [27]. Even when technical aspects of the platform run flawlessly, biologic errors can still occur. One source of biologic error is related to the presence of chromosomal mosaicism within the developing embryo, which is characterized by the presence of a mixture of diploid and aneuploid cell lines. Mosaic embryos may not transferred usually because they are deemed abnormal. However, single TE biopsy cannot reliably reflect the whole TE and ICM [28]. Moreover, the ability of aCGH to detect mosaicism is dependent on the percentage of aneuploid cells in the TE biopsy. NGS now has the ability to detect mosaicism molecularly. The higher rates of reporting mosaicism by NGS compared with aCGH have called into question the validity of a diagnosis of mosaicism [29]. So if some viable embryos are discarded either because of false positives or mosaicism, then for DOR patients, where the number of available oocytes and embryos is strictly limited, treatment may be compromised [30].
Furthermore, the implementation of expensive technologies in healthcare, even when shown to offer an incremental improvement to outcomes, requires careful consideration of their relative cost-effectiveness. Currently, the cost of PGT-A remains high, making it unaffordable in many patients if not paid for by health insurance. In addition, it is difficult to quantify the intangible costs of failed implantation, miscarriage, all obstetric, neonatal, and ongoing costs of disease/aneuploidy. More research is needed to help patients, clinicians, and insurers in their decision regarding whether to utilize and pay for this technology [22].
Recently after reviewing the data, the American Society of Reproductive Medicine (ASRM) concluded that studies on use of PGT-A have limitations, and there are concerns about appropriate patient selections and testing platforms. For instance, patients in RCTs are mostly good responders and some started randomization on the day of blastulation rather than on the day of commencing stimulation. The questions about false-positive testing, potential embryonic damage with biopsy, loss of potential euploid embryos between day 3 and day 5, and blastocyst formation are not addressed. As we also discussed in Chap. 19, ASRM also agrees that not all the embryos survive in culture to develop into blastocyst stage for biopsy, although potentially they may have resulted in live birth if transferred in the cleavage stage [22]. Debates continue on the pros and cons of PGT-A [31].
24.6 Conclusion
The techniques of PGT-A, blastocyst culture, and biopsy as well as freeze-all approaches have the potential to reduce time to ongoing pregnancy and decrease miscarriages and ongoing aneuploid gestations in DOR patients. A complete assessment of the efficacy in this population will require more information regarding genetic variations, the biological ovarian aging, and multiple phenotypes of DOR. At present, however, there is insufficient evidence to recommend the routine use of blastocyst biopsy for aneuploidy testing in DOR patients. Therefore, taking into consideration each patient’s unique circumstance, extensive counseling based on the pros and cons of the PGT-A procedure should be provided to best take care of this challenging patient population.

Full access? Get Clinical Tree


